EDS_Tools: Spectroscopy
Analysis of EDS Spectrum Images¶

part of
a pycroscopy ecosystem package
Notebook by Gerd Duscher, 2025
Microscopy Facilities
Institute of Advanced Materials & Manufacturing
The University of Tennessee, Knoxville
Model based analysis and quantification of data acquired with transmission electron microscopes
Content¶
An Introduction into displaying and analyzing EDS spectrum images and spectra This works also on Google Colab.
Unlike in an EELS spectrum, a single pixel in an EDS spectrum image does not contain enough information to quantify the chemical composition or even detect trace elements.
Several different stategies will be explored in this notebook of how to analyze these kind of data.
Prerequesites¶
Install pyTEMlib¶
If you have not done so in the Introduction Notebook, please test and install pyTEMlib and other important packages with the code cell below.
import sys
import importlib.metadata
def test_package(package_name):
"""Test if package exists and returns version or -1"""
try:
version = importlib.metadata.version(package_name)
except importlib.metadata.PackageNotFoundError:
version = '-1'
return version
# pyTEMlib setup ------------------
if test_package('pyTEMlib') < '0.2025.12.0':
print('installing pyTEMlib')
!{sys.executable} -m pip install pyTEMlib --upgrade
# ------------------------------
print('done')installing pyTEMlib
Requirement already satisfied: pyTEMlib in c:\users\gduscher\appdata\local\anaconda3\lib\site-packages (0.2025.11.3)
Collecting pyTEMlib
Downloading pytemlib-0.2025.12.0-py3-none-any.whl.metadata (3.6 kB)
Requirement already satisfied: scipy in c:\users\gduscher\appdata\local\anaconda3\lib\site-packages (from pyTEMlib) (1.16.3)
Requirement already satisfied: numpy in c:\users\gduscher\appdata\local\anaconda3\lib\site-packages (from pyTEMlib) (2.3.5)
Requirement already satisfied: pillow in c:\users\gduscher\appdata\local\anaconda3\lib\site-packages (from pyTEMlib) (12.0.0)
Requirement already satisfied: ase in c:\users\gduscher\appdata\local\anaconda3\lib\site-packages (from pyTEMlib) (3.26.0)
Requirement already satisfied: tqdm in c:\users\gduscher\appdata\local\anaconda3\lib\site-packages (from pyTEMlib) (4.67.1)
Requirement already satisfied: plotly in c:\users\gduscher\appdata\local\anaconda3\lib\site-packages (from pyTEMlib) (6.5.0)
Requirement already satisfied: pandas in c:\users\gduscher\appdata\local\anaconda3\lib\site-packages (from pyTEMlib) (2.3.3)
Requirement already satisfied: requests in c:\users\gduscher\appdata\local\anaconda3\lib\site-packages (from pyTEMlib) (2.32.5)
Requirement already satisfied: lxml in c:\users\gduscher\appdata\local\anaconda3\lib\site-packages (from pyTEMlib) (5.3.0)
Requirement already satisfied: ipympl in c:\users\gduscher\appdata\local\anaconda3\lib\site-packages (from pyTEMlib) (0.9.7)
Requirement already satisfied: spglib in c:\users\gduscher\appdata\local\anaconda3\lib\site-packages (from pyTEMlib) (2.6.0)
Requirement already satisfied: simpleitk in c:\users\gduscher\appdata\local\anaconda3\lib\site-packages (from pyTEMlib) (2.5.2)
Requirement already satisfied: scikit-image in c:\users\gduscher\appdata\local\anaconda3\lib\site-packages (from pyTEMlib) (0.25.2)
Requirement already satisfied: scikit-learn in c:\users\gduscher\appdata\local\anaconda3\lib\site-packages (from pyTEMlib) (1.7.2)
Requirement already satisfied: pyNSID>=0.0.7 in c:\users\gduscher\appdata\local\anaconda3\lib\site-packages (from pyTEMlib) (0.0.7.2)
Requirement already satisfied: sidpy>=0.12.7 in c:\users\gduscher\appdata\local\anaconda3\lib\site-packages (from pyTEMlib) (0.12.7)
Requirement already satisfied: SciFiReaders>=0.12.4 in c:\users\gduscher\appdata\local\anaconda3\lib\site-packages (from pyTEMlib) (0.12.4)
Requirement already satisfied: xraylib in c:\users\gduscher\appdata\local\anaconda3\lib\site-packages (from pyTEMlib) (4.2.0)
Requirement already satisfied: toolz in c:\users\gduscher\appdata\local\anaconda3\lib\site-packages (from pyNSID>=0.0.7->pyTEMlib) (1.0.0)
Requirement already satisfied: cytoolz in c:\users\gduscher\appdata\local\anaconda3\lib\site-packages (from pyNSID>=0.0.7->pyTEMlib) (1.0.1)
Requirement already satisfied: dask>=0.10 in c:\users\gduscher\appdata\local\anaconda3\lib\site-packages (from pyNSID>=0.0.7->pyTEMlib) (2025.11.0)
Requirement already satisfied: h5py>=2.6.0 in c:\users\gduscher\appdata\local\anaconda3\lib\site-packages (from pyNSID>=0.0.7->pyTEMlib) (3.15.1)
Requirement already satisfied: six in c:\users\gduscher\appdata\local\anaconda3\lib\site-packages (from pyNSID>=0.0.7->pyTEMlib) (1.17.0)
Requirement already satisfied: click>=8.1 in c:\users\gduscher\appdata\local\anaconda3\lib\site-packages (from dask>=0.10->pyNSID>=0.0.7->pyTEMlib) (8.1.8)
Requirement already satisfied: cloudpickle>=3.0.0 in c:\users\gduscher\appdata\local\anaconda3\lib\site-packages (from dask>=0.10->pyNSID>=0.0.7->pyTEMlib) (3.1.1)
Requirement already satisfied: fsspec>=2021.09.0 in c:\users\gduscher\appdata\local\anaconda3\lib\site-packages (from dask>=0.10->pyNSID>=0.0.7->pyTEMlib) (2025.10.0)
Requirement already satisfied: packaging>=20.0 in c:\users\gduscher\appdata\local\anaconda3\lib\site-packages (from dask>=0.10->pyNSID>=0.0.7->pyTEMlib) (25.0)
Requirement already satisfied: partd>=1.4.0 in c:\users\gduscher\appdata\local\anaconda3\lib\site-packages (from dask>=0.10->pyNSID>=0.0.7->pyTEMlib) (1.4.2)
Requirement already satisfied: pyyaml>=5.3.1 in c:\users\gduscher\appdata\local\anaconda3\lib\site-packages (from dask>=0.10->pyNSID>=0.0.7->pyTEMlib) (6.0.3)
Requirement already satisfied: colorama in c:\users\gduscher\appdata\local\anaconda3\lib\site-packages (from click>=8.1->dask>=0.10->pyNSID>=0.0.7->pyTEMlib) (0.4.6)
Requirement already satisfied: locket in c:\users\gduscher\appdata\local\anaconda3\lib\site-packages (from partd>=1.4.0->dask>=0.10->pyNSID>=0.0.7->pyTEMlib) (1.0.0)
Requirement already satisfied: setuptools in c:\users\gduscher\appdata\local\anaconda3\lib\site-packages (from SciFiReaders>=0.12.4->pyTEMlib) (80.9.0)
Requirement already satisfied: numba in c:\users\gduscher\appdata\local\anaconda3\lib\site-packages (from SciFiReaders>=0.12.4->pyTEMlib) (0.62.1)
Requirement already satisfied: ipython>=7.1.0 in c:\users\gduscher\appdata\local\anaconda3\lib\site-packages (from SciFiReaders>=0.12.4->pyTEMlib) (9.7.0)
Requirement already satisfied: pyUSID in c:\users\gduscher\appdata\local\anaconda3\lib\site-packages (from SciFiReaders>=0.12.4->pyTEMlib) (0.0.12)
Requirement already satisfied: gdown in c:\users\gduscher\appdata\local\anaconda3\lib\site-packages (from SciFiReaders>=0.12.4->pyTEMlib) (5.2.0)
Requirement already satisfied: mrcfile in c:\users\gduscher\appdata\local\anaconda3\lib\site-packages (from SciFiReaders>=0.12.4->pyTEMlib) (1.5.4)
Requirement already satisfied: pycroscopy-gwyfile in c:\users\gduscher\appdata\local\anaconda3\lib\site-packages (from SciFiReaders>=0.12.4->pyTEMlib) (1.0.2)
Requirement already satisfied: decorator>=4.3.2 in c:\users\gduscher\appdata\local\anaconda3\lib\site-packages (from ipython>=7.1.0->SciFiReaders>=0.12.4->pyTEMlib) (5.2.1)
Requirement already satisfied: ipython-pygments-lexers>=1.0.0 in c:\users\gduscher\appdata\local\anaconda3\lib\site-packages (from ipython>=7.1.0->SciFiReaders>=0.12.4->pyTEMlib) (1.1.1)
Requirement already satisfied: jedi>=0.18.1 in c:\users\gduscher\appdata\local\anaconda3\lib\site-packages (from ipython>=7.1.0->SciFiReaders>=0.12.4->pyTEMlib) (0.19.2)
Requirement already satisfied: matplotlib-inline>=0.1.5 in c:\users\gduscher\appdata\local\anaconda3\lib\site-packages (from ipython>=7.1.0->SciFiReaders>=0.12.4->pyTEMlib) (0.2.1)
Requirement already satisfied: prompt_toolkit<3.1.0,>=3.0.41 in c:\users\gduscher\appdata\local\anaconda3\lib\site-packages (from ipython>=7.1.0->SciFiReaders>=0.12.4->pyTEMlib) (3.0.52)
Requirement already satisfied: pygments>=2.11.0 in c:\users\gduscher\appdata\local\anaconda3\lib\site-packages (from ipython>=7.1.0->SciFiReaders>=0.12.4->pyTEMlib) (2.19.2)
Requirement already satisfied: stack_data>=0.6.0 in c:\users\gduscher\appdata\local\anaconda3\lib\site-packages (from ipython>=7.1.0->SciFiReaders>=0.12.4->pyTEMlib) (0.6.3)
Requirement already satisfied: traitlets>=5.13.0 in c:\users\gduscher\appdata\local\anaconda3\lib\site-packages (from ipython>=7.1.0->SciFiReaders>=0.12.4->pyTEMlib) (5.14.3)
Requirement already satisfied: wcwidth in c:\users\gduscher\appdata\local\anaconda3\lib\site-packages (from prompt_toolkit<3.1.0,>=3.0.41->ipython>=7.1.0->SciFiReaders>=0.12.4->pyTEMlib) (0.2.13)
Requirement already satisfied: parso<0.9.0,>=0.8.4 in c:\users\gduscher\appdata\local\anaconda3\lib\site-packages (from jedi>=0.18.1->ipython>=7.1.0->SciFiReaders>=0.12.4->pyTEMlib) (0.8.5)
Requirement already satisfied: matplotlib>=2.0.0 in c:\users\gduscher\appdata\local\anaconda3\lib\site-packages (from sidpy>=0.12.7->pyTEMlib) (3.10.6)
Requirement already satisfied: distributed>=2.0.0 in c:\users\gduscher\appdata\local\anaconda3\lib\site-packages (from sidpy>=0.12.7->pyTEMlib) (2025.11.0)
Requirement already satisfied: psutil in c:\users\gduscher\appdata\local\anaconda3\lib\site-packages (from sidpy>=0.12.7->pyTEMlib) (7.0.0)
Requirement already satisfied: joblib>=0.11.0 in c:\users\gduscher\appdata\local\anaconda3\lib\site-packages (from sidpy>=0.12.7->pyTEMlib) (1.5.2)
Requirement already satisfied: ipywidgets in c:\users\gduscher\appdata\local\anaconda3\lib\site-packages (from sidpy>=0.12.7->pyTEMlib) (8.1.7)
Requirement already satisfied: ipykernel in c:\users\gduscher\appdata\local\anaconda3\lib\site-packages (from sidpy>=0.12.7->pyTEMlib) (6.31.0)
Requirement already satisfied: dill in c:\users\gduscher\appdata\local\anaconda3\lib\site-packages (from sidpy>=0.12.7->pyTEMlib) (0.4.0)
Requirement already satisfied: jinja2>=2.10.3 in c:\users\gduscher\appdata\local\anaconda3\lib\site-packages (from distributed>=2.0.0->sidpy>=0.12.7->pyTEMlib) (3.1.6)
Requirement already satisfied: msgpack>=1.0.2 in c:\users\gduscher\appdata\local\anaconda3\lib\site-packages (from distributed>=2.0.0->sidpy>=0.12.7->pyTEMlib) (1.1.1)
Requirement already satisfied: sortedcontainers>=2.0.5 in c:\users\gduscher\appdata\local\anaconda3\lib\site-packages (from distributed>=2.0.0->sidpy>=0.12.7->pyTEMlib) (2.4.0)
Requirement already satisfied: tblib>=1.6.0 in c:\users\gduscher\appdata\local\anaconda3\lib\site-packages (from distributed>=2.0.0->sidpy>=0.12.7->pyTEMlib) (3.1.0)
Requirement already satisfied: tornado>=6.2.0 in c:\users\gduscher\appdata\local\anaconda3\lib\site-packages (from distributed>=2.0.0->sidpy>=0.12.7->pyTEMlib) (6.5.1)
Requirement already satisfied: urllib3>=1.26.5 in c:\users\gduscher\appdata\local\anaconda3\lib\site-packages (from distributed>=2.0.0->sidpy>=0.12.7->pyTEMlib) (2.5.0)
Requirement already satisfied: zict>=3.0.0 in c:\users\gduscher\appdata\local\anaconda3\lib\site-packages (from distributed>=2.0.0->sidpy>=0.12.7->pyTEMlib) (3.0.0)
Requirement already satisfied: MarkupSafe>=2.0 in c:\users\gduscher\appdata\local\anaconda3\lib\site-packages (from jinja2>=2.10.3->distributed>=2.0.0->sidpy>=0.12.7->pyTEMlib) (3.0.2)
Requirement already satisfied: contourpy>=1.0.1 in c:\users\gduscher\appdata\local\anaconda3\lib\site-packages (from matplotlib>=2.0.0->sidpy>=0.12.7->pyTEMlib) (1.3.3)
Requirement already satisfied: cycler>=0.10 in c:\users\gduscher\appdata\local\anaconda3\lib\site-packages (from matplotlib>=2.0.0->sidpy>=0.12.7->pyTEMlib) (0.11.0)
Requirement already satisfied: fonttools>=4.22.0 in c:\users\gduscher\appdata\local\anaconda3\lib\site-packages (from matplotlib>=2.0.0->sidpy>=0.12.7->pyTEMlib) (4.60.1)
Requirement already satisfied: kiwisolver>=1.3.1 in c:\users\gduscher\appdata\local\anaconda3\lib\site-packages (from matplotlib>=2.0.0->sidpy>=0.12.7->pyTEMlib) (1.4.9)
Requirement already satisfied: pyparsing>=2.3.1 in c:\users\gduscher\appdata\local\anaconda3\lib\site-packages (from matplotlib>=2.0.0->sidpy>=0.12.7->pyTEMlib) (3.2.5)
Requirement already satisfied: python-dateutil>=2.7 in c:\users\gduscher\appdata\local\anaconda3\lib\site-packages (from matplotlib>=2.0.0->sidpy>=0.12.7->pyTEMlib) (2.9.0.post0)
Requirement already satisfied: executing>=1.2.0 in c:\users\gduscher\appdata\local\anaconda3\lib\site-packages (from stack_data>=0.6.0->ipython>=7.1.0->SciFiReaders>=0.12.4->pyTEMlib) (2.2.1)
Requirement already satisfied: asttokens>=2.1.0 in c:\users\gduscher\appdata\local\anaconda3\lib\site-packages (from stack_data>=0.6.0->ipython>=7.1.0->SciFiReaders>=0.12.4->pyTEMlib) (3.0.0)
Requirement already satisfied: pure_eval in c:\users\gduscher\appdata\local\anaconda3\lib\site-packages (from stack_data>=0.6.0->ipython>=7.1.0->SciFiReaders>=0.12.4->pyTEMlib) (0.2.3)
Requirement already satisfied: beautifulsoup4 in c:\users\gduscher\appdata\local\anaconda3\lib\site-packages (from gdown->SciFiReaders>=0.12.4->pyTEMlib) (4.14.2)
Requirement already satisfied: filelock in c:\users\gduscher\appdata\local\anaconda3\lib\site-packages (from gdown->SciFiReaders>=0.12.4->pyTEMlib) (3.20.0)
Requirement already satisfied: soupsieve>1.2 in c:\users\gduscher\appdata\local\anaconda3\lib\site-packages (from beautifulsoup4->gdown->SciFiReaders>=0.12.4->pyTEMlib) (2.5)
Requirement already satisfied: typing-extensions>=4.0.0 in c:\users\gduscher\appdata\local\anaconda3\lib\site-packages (from beautifulsoup4->gdown->SciFiReaders>=0.12.4->pyTEMlib) (4.15.0)
Requirement already satisfied: comm>=0.1.1 in c:\users\gduscher\appdata\local\anaconda3\lib\site-packages (from ipykernel->sidpy>=0.12.7->pyTEMlib) (0.2.3)
Requirement already satisfied: debugpy>=1.6.5 in c:\users\gduscher\appdata\local\anaconda3\lib\site-packages (from ipykernel->sidpy>=0.12.7->pyTEMlib) (1.8.16)
Requirement already satisfied: jupyter-client>=8.0.0 in c:\users\gduscher\appdata\local\anaconda3\lib\site-packages (from ipykernel->sidpy>=0.12.7->pyTEMlib) (8.6.3)
Requirement already satisfied: jupyter-core!=5.0.*,>=4.12 in c:\users\gduscher\appdata\local\anaconda3\lib\site-packages (from ipykernel->sidpy>=0.12.7->pyTEMlib) (5.9.1)
Requirement already satisfied: nest-asyncio>=1.4 in c:\users\gduscher\appdata\local\anaconda3\lib\site-packages (from ipykernel->sidpy>=0.12.7->pyTEMlib) (1.6.0)
Requirement already satisfied: pyzmq>=25 in c:\users\gduscher\appdata\local\anaconda3\lib\site-packages (from ipykernel->sidpy>=0.12.7->pyTEMlib) (27.1.0)
Requirement already satisfied: platformdirs>=2.5 in c:\users\gduscher\appdata\local\anaconda3\lib\site-packages (from jupyter-core!=5.0.*,>=4.12->ipykernel->sidpy>=0.12.7->pyTEMlib) (4.5.0)
Requirement already satisfied: widgetsnbextension~=4.0.14 in c:\users\gduscher\appdata\local\anaconda3\lib\site-packages (from ipywidgets->sidpy>=0.12.7->pyTEMlib) (4.0.14)
Requirement already satisfied: jupyterlab_widgets~=3.0.15 in c:\users\gduscher\appdata\local\anaconda3\lib\site-packages (from ipywidgets->sidpy>=0.12.7->pyTEMlib) (3.0.15)
Requirement already satisfied: llvmlite<0.46,>=0.45.0dev0 in c:\users\gduscher\appdata\local\anaconda3\lib\site-packages (from numba->SciFiReaders>=0.12.4->pyTEMlib) (0.45.1)
Requirement already satisfied: pytz>=2020.1 in c:\users\gduscher\appdata\local\anaconda3\lib\site-packages (from pandas->pyTEMlib) (2025.2)
Requirement already satisfied: tzdata>=2022.7 in c:\users\gduscher\appdata\local\anaconda3\lib\site-packages (from pandas->pyTEMlib) (2025.2)
Requirement already satisfied: narwhals>=1.15.1 in c:\users\gduscher\appdata\local\anaconda3\lib\site-packages (from plotly->pyTEMlib) (2.7.0)
Requirement already satisfied: charset_normalizer<4,>=2 in c:\users\gduscher\appdata\local\anaconda3\lib\site-packages (from requests->pyTEMlib) (3.4.4)
Requirement already satisfied: idna<4,>=2.5 in c:\users\gduscher\appdata\local\anaconda3\lib\site-packages (from requests->pyTEMlib) (3.11)
Requirement already satisfied: certifi>=2017.4.17 in c:\users\gduscher\appdata\local\anaconda3\lib\site-packages (from requests->pyTEMlib) (2025.11.12)
Requirement already satisfied: PySocks!=1.5.7,>=1.5.6 in c:\users\gduscher\appdata\local\anaconda3\lib\site-packages (from requests[socks]->gdown->SciFiReaders>=0.12.4->pyTEMlib) (1.7.1)
Requirement already satisfied: networkx>=3.0 in c:\users\gduscher\appdata\local\anaconda3\lib\site-packages (from scikit-image->pyTEMlib) (3.5)
Requirement already satisfied: imageio!=2.35.0,>=2.33 in c:\users\gduscher\appdata\local\anaconda3\lib\site-packages (from scikit-image->pyTEMlib) (2.37.2)
Requirement already satisfied: tifffile>=2022.8.12 in c:\users\gduscher\appdata\local\anaconda3\lib\site-packages (from scikit-image->pyTEMlib) (2025.10.4)
Requirement already satisfied: lazy-loader>=0.4 in c:\users\gduscher\appdata\local\anaconda3\lib\site-packages (from scikit-image->pyTEMlib) (0.4)
Requirement already satisfied: threadpoolctl>=3.1.0 in c:\users\gduscher\appdata\local\anaconda3\lib\site-packages (from scikit-learn->pyTEMlib) (3.5.0)
Downloading pytemlib-0.2025.12.0-py3-none-any.whl (1.5 MB)
---------------------------------------- 0.0/1.5 MB ? eta -:--:--
----------------------------------- ---- 1.3/1.5 MB 13.1 MB/s eta 0:00:01
---------------------------------------- 1.5/1.5 MB 8.2 MB/s 0:00:00
Installing collected packages: pyTEMlib
Attempting uninstall: pyTEMlib
Found existing installation: pyTEMlib 0.2025.11.3
Uninstalling pyTEMlib-0.2025.11.3:
Successfully uninstalled pyTEMlib-0.2025.11.3
Successfully installed pyTEMlib-0.2025.12.0
done
WARNING: The script pyTEMlib.exe is installed in 'c:\Users\gduscher\AppData\Local\anaconda3\Scripts' which is not on PATH.
Consider adding this directory to PATH or, if you prefer to suppress this warning, use --no-warn-script-location.
ERROR: pip's dependency resolver does not currently take into account all the packages that are installed. This behaviour is the source of the following dependency conflicts.
pycrosgui 0.2.2 requires pyqt5, which is not installed.
pytemgui 0.2.2 requires pyqt5, which is not installed.
Loading of necessary libraries¶
Please note, that we only need to load the pyTEMlib library, which is based on sidpy Datsets.
%matplotlib widget
import sys
import numpy as np
import matplotlib.pylab as plt
# using pyTEMlib.eds_tools, pyTEMlib.file_tools and pyTEMlib.eels_tools (for line definitions)
%load_ext autoreload
%autoreload 2
sys.path.insert(0, '../../')
import pyTEMlib
if 'google.colab' in sys.modules:
from google.colab import output
output.enable_custom_widget_manager()
from google.colab import drive
if 'google.colab' in sys.modules:
drive.mount("/content/drive")
# For archiving reasons it is a good idea to print the version numbers out at this point
print('pyTEM version: ',pyTEMlib.__version__)
__notebook__ = 'EDS_Spectrum_Analysis'
__notebook_version__ = '2025_10_27'pyTEM version: 0.2026.1.3
Open File¶
Load File¶
Select a main dataset and any additional data like reference data and such. Go to open and plot dataset for more explanations ()
#path = "C:\Users\gduscher\OneDrive - University of Tennessee\google_drive\2022 Experiments\Spectra\20221214\AlCe-200kV"
fileWidget = pyTEMlib.file_tools.FileWidget()dataset = fileWidget.selected_dataset
view = dataset.plot()
print(dataset)
datasetsidpy.Dataset of type IMAGE with:
dask.array<array, shape=(268, 222), dtype=float32, chunksize=(268, 222), chunktype=numpy.ndarray>
data contains: intensity (counts)
and Dimensions:
x: distance (nm) of size (268,)
y: distance (nm) of size (222,)
with metadata: ['experiment', 'EDS', 'filename']
import scipy
smoo = scipy.ndimage.gaussian_filter(dataset, 7)
plt.figure()
plt.imshow(smoo.T)
plt.colorbar().1/dataset.x.slope
np.float64(13.313608508711992)Clicking on the image will show the coordinates of the pixel clicked and displays that spectrum.
One sees that the spectra contain too little counts for a meaningful analysis. To improve the statistics we will sum up spectra from a region of interest (ROI).
Different stategies for this summation will be explored below.
Rigid Registration and Atom Finding¶
Select the Image stack
chooser = pyTEMlib.file_tools.ChooseDataset(fileWidget.datasets)haadf = chooser.dataset
rigid_registered = pyTEMlib.image_tools.rigid_registration(haadf)
v = rigid_registered.plot()Visualize Drift
drift = rigid_registered.metadata['analysis']['rigid_registration']['drift']
polynom_degree = 2 # 1 is linear fit, 2 is parabolic fit, ...
x = np.linspace(0,drift.shape[0]-1,drift.shape[0])
line_fit_x = np.polyfit(x, drift[:,0], polynom_degree)
poly_x = np.poly1d(line_fit_x)
line_fit_y = np.polyfit(x, drift[:,1], polynom_degree)
poly_y = np.poly1d(line_fit_y)
plt.figure()
plt.axhline(color = 'gray')
plt.plot(x, drift[:,0], label = 'drift x')
plt.plot(x, drift[:,1], label = 'drift y')
plt.plot(x, poly_x(x), label = 'fit_drift_x')
plt.plot(x, poly_y(x), label = 'fit_drift_y')
plt.legend();
ax_pixels = plt.gca()
ax_pixels.step(1, 1)
scaleX = (rigid_registered.x[1]-rigid_registered.x[0])*1000. #in pm
ax_pm = ax_pixels.twinx()
x_1, x_2 = ax_pixels.get_ylim()
ax_pm.set_ylim(x_1*scaleX, x_2*scaleX)
ax_pixels.set_ylabel('drift [pixels]')
ax_pm.set_ylabel('drift [pm]')
ax_pixels.set_xlabel('image number');
plt.tight_layout()haadf_sum = haadf.sum(axis=0)
haadf_sum.data_type = 'image'
plt.figure()
plt.imshow(haadf_sum.T, cmap='gray');width = 7
position = np.array([67,136])-int(width/2)
position = np.array([127,98])-int(width/2)
atom = haadf_sum[position[0]:position[0]+width, position[1]:position[1]+width]
atom.data_type = 'image'
atom.plot();chooser = pyTEMlib.file_tools.ChooseDataset(fileWidget.datasets)dataset = chooser.dataset
atom_sum = dataset[position[0]:position[0]+width, position[1]:position[1]+width]
atom_sum= atom_sum.mean(axis=(0,1))
spectrum_sum = dataset.mean(axis=(0,1))
spectrum_sum.data_type = 'spectrum'
view = spectrum_sum.plot();
#plt.gca().plot(spectrum_sum.energy_scale.values, np.array(dataset[118, 81])/10, label='single pixel');
#plt.gca().plot(spectrum_sum.energy_scale.values, atom_sum, label='atom')
plt.gca().plot(spectrum_sum.energy_scale.values, spectrum_sum, label='sum')
plt.gca().plot(spectrum_sum.energy_scale.values, scipy.ndimage.gaussian_filter(atom_sum-spectrum_sum, 2), label='difference')
plt.gca().legend();atom_sum = dataset[position[0]:position[0]+width, position[1]:position[1]+width]
atom_sum= atom_sum.mean(axis=(0,1))
atom_sum.data_type = 'Spectrum'
atom_sum.plot()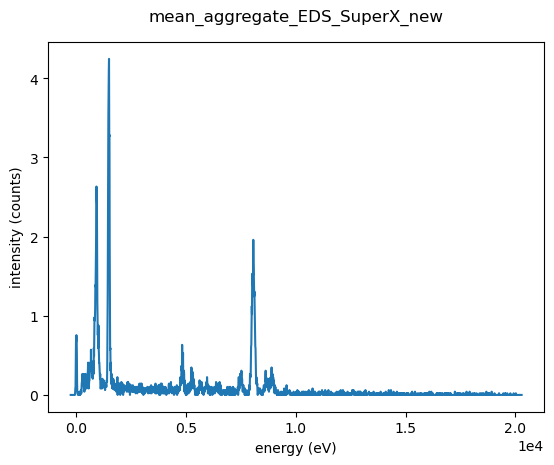
This is a clear demonstration that even the sum over a single atom will not give a good statistic in the EDS spectra
dataset.view_metadata()experiment :
detector : SuperXG21
acceleration_voltage : 200000.0
microscope : Titan
start_date_time : 1671044532
collection_angle : -1.0
convergence_angle : 0.03
probe_mode : convergent
stage :
holder :
position :
x : -0.0004458012750000001
y : 0.00041000946000000014
z : -5.289224220235313e-05
tilt :
alpha : 0.12116822300000002
beta : 0.08483282720148563
instrument : Spectra4018
current : 6.9062810780693e-11
pixel_time : 5e-05
exposure_time : 2.2339
collection_angle_end : -1.0
EDS :
detector :
layers :
13 :
thickness : 5e-08
Z : 13
element : Al
SiDeadThickness : 1.3e-07
SiLiveThickness : 0.05
detector_area : 2.9999999999999997e-05
energy_resolution : 125
start_energy : 120
start_channel : 100
ElevationAngle : 0.31415927
AzimuthAngle : 0.7853981633974483
RealTime : 1.927908375
LiveTime : 1.912787525
filename : C:\Users\gduscher\OneDrive - University of Tennessee\google_drive\2022 Experiments\Spectra\20221214\AlCe-200kV\SI HAADF 1402.emd
### Does not work for spectrum images
#
spectrum = dataset.sum(axis=(0,1))
spectrum.data_type = 'spectrum'
start = np.searchsorted(spectrum.energy_scale.values, 100)
energy_scale = spectrum.energy_scale.values[start:]
detector_Efficiency= pyTEMlib.eds_tools.detector_response(spectrum) # tags, spectrum.energy_scale.values[start:])
if 'start_energy' not in spectrum.metadata['EDS']['detector']:
spectrum.metadata['EDS']['detector']['start_energy'] = 120
spectrum[:np.searchsorted(spectrum.energy_scale.values,spectrum.metadata['EDS']['detector']['start_energy'])] = 0.
spectrum.metadata['EDS']['detector']['detector_efficiency'] = detector_EfficiencyFind Elements¶
# --------Input -----------
minimum_number_of_peaks = 10
# --------------------------
import scipy
spectrum = atom_sum.copy()
pyTEMlib.eds_tools.get_elements(spectrum, minimum_number_of_peaks, verbose=False)
#atom_sum = dataset[position[0]:position[0]+width, position[1]:position[1]+width]
#atom_sum= atom_sum.sum(axis=(0,1))
# atom_sum = scipy.ndimage.gaussian_filter1d(atom_sum, sigma=3)
#single = dataset[87,84]
plt.figure()
plt.plot(spectrum.energy_scale,spectrum, label = 'spectrum')
pyTEMlib.eds_tools.plot_lines(spectrum.metadata['EDS'], plt.gca())
#plt.gca().plot(spectrum_sum.energy_scale.values, np.array(single)*300, label='single pixel');
#plt.gca().plot(spectrum_sum.energy_scale.values, atom_sum*100)
#plt.gca().plot(spectrum_sum.energy_scale.values, atom_sum*100, label='atom')
plt.gca().plot(spectrum_sum.energy_scale.values, spectrum, label='sum')
plt.gca().plot(spectrum_sum.energy_scale.values, scipy.ndimage.gaussian_filter(atom_sum-spectrum_sum, 3), label='difference')
plt.gca().legend();C:\Users\gduscher\AppData\Local\anaconda3\Lib\site-packages\dask\array\core.py:1738: FutureWarning: The `numpy.argsort` function is not implemented by Dask array. You may want to use the da.map_blocks function or something similar to silence this warning. Your code may stop working in a future release.
warnings.warn(
spectrum_sum.metadata['EDS'].keys()dict_keys(['detector', 'bremsstrahlung', 'GUI', 'Ni', 'S', 'Cu', 'Ce', 'Nd', 'O', 'C', 'Al', 'Zn', 'F'])del spectrum_sum.metadata['EDS']['O']spectrum_sum.metadata['EDS'].keys()
#del spectrum_sum.metadata['EDS']['Co']
plt.figure()
plt.plot(spectrum.energy_scale,spectrum, label = 'spectrum')
pyTEMlib.eds_tools.plot_lines(spectrum.metadata['EDS'], plt.gca())
#plt.gca().plot(spectrum_sum.energy_scale.values, np.array(single)*300, label='single pixel');
#plt.gca().plot(spectrum_sum.energy_scale.values, atom_sum*100)
#plt.gca().plot(spectrum_sum.energy_scale.values, atom_sum*100, label='atom')
plt.gca().plot(spectrum_sum.energy_scale.values, spectrum, label='sum')
plt.gca().plot(spectrum_sum.energy_scale.values, scipy.ndimage.gaussian_filter(atom_sum-spectrum_sum, 3), label='difference')
plt.gca().legend();print(spectrum)sidpy.Dataset of type SPECTRUM with:
dask.array<mean_agg-aggregate, shape=(2048,), dtype=float64, chunksize=(2048,), chunktype=numpy.ndarray>
data contains: intensity (counts)
and Dimensions:
energy_scale: energy (eV) of size (2048,)
with metadata: ['experiment', 'EDS', 'filename']
### Add Mg
def add_element(spectrum, elements):
if isinstance(elements, str):
elements = [elements]
if not isinstance(elements, list):
raise ValueError("elements must be a string or a list of strings")
spectrum.metadata['EDS'].update(pyTEMlib.eds_tools.get_x_ray_lines(spectrum, elements))
add_element(spectrum, 'Mn')
spectrum.metadata['EDS']['Mg'] {'Z': 12,
'K-family': {'lines': ['K-L3', 'K-L2'],
'main': 'K-L3',
'weight': 1.0,
'height': np.float64(0.1836734693877551),
'K-L3': {'weight': 1.0, 'position': 1253.6},
'K-L2': {'weight': 0.507, 'position': 1253.6},
'peaks': array([0., 0., 0., ..., 0., 0., 0.], shape=(2048,)),
'areal_density': np.float64(0.35894005285976044),
'fluorescent_yield': 0.02333},
'atomic_weight': 24.305,
'nominal_density': 1.735}out_string = 'Found Elements: '
for key in spectrum.metadata['EDS']:
if len(key) <3:
out_string += f'{key}, '
print(out_string)Found Elements: Ni, Cu, Ce, Al, Zn, Mg,
Viziualization what happens at the atomic scale.
We plot the difference between the atom signal and the total signal of the spectrum-image
atom_sum = dataset[position[0]:position[0]+width, position[1]:position[1]+width]
atom_sum= atom_sum.sum(axis=(0,1))
atom_sum = scipy.ndimage.gaussian_filter1d(atom_sum, sigma=3)
spectrum_ref = spectrum/spectrum.sum()*atom_sum.sum()
spectrum_ref = scipy.ndimage.gaussian_filter1d(spectrum_ref, sigma=3)
difference = atom_sum - spectrum_ref
difference = difference -np.mean(difference)
#difference[difference < 0] = 0
difference = scipy.ndimage.gaussian_filter1d(difference, sigma=3)
plt.figure()
plt.plot(spectrum.energy_scale,difference*1000, label = 'spectrum')
pyTEMlib.eds_tools.plot_lines(spectrum.metadata['EDS'], plt.gca())
plt.axhline(0, color='gray')
spectrum = spectrum_sum#dataset.sum(axis=(0,1))
spectrum.data_type = 'spectrum'
atom_sum = dataset[position[0]:position[0]+width, position[1]:position[1]+width]
atom_sum = atom_sum.sum(axis=(0,1))
atom_sum = scipy.ndimage.gaussian_filter1d(atom_sum, sigma=3)
spectrum_atom = spectrum.like_data(atom_sum).copy()
spectrum_atom.data_type = 'spectrum'import pyTEMlib.eds_tools
spectrum = atom_sum
peaks, pp = pyTEMlib.eds_tools.fit_model(spectrum, use_detector_efficiency=True)
model = pyTEMlib.eds_tools.get_model(spectrum)
plt.figure()
plt.plot(spectrum.energy_scale, spectrum, label='spectrum')
plt.plot(spectrum.energy_scale, model, label='model')
plt.plot(spectrum.energy_scale, np.array(spectrum-model), label='difference')
plt.xlabel('energy (eV)')
pyTEMlib.eds_tools.plot_lines(spectrum.metadata['EDS'], plt.gca())
plt.axhline(y=0, xmin=0, xmax=1, color='gray')
plt.legend()no intensity Ce M-family
Quantify Spectrum¶
first with Bote-Salvat cross section using dictionaries calculated with emtables package.
pyTEMlib.eds_tools.quantify_eds(atom_sum, mask=[])using cross sections for quantification
Ni: 4.22 at% 4.87 wt%
Cu: 37.55 at% 46.93 wt%
Ce: 6.17 at% 17.01 wt%
Al: 46.39 at% 24.61 wt%
Zn: 3.66 at% 4.71 wt%
Mn: 1.51 at% 1.64 wt%
Mg: 0.50 at% 0.24 wt%
then with k-factor dictionary
print('whole spectrum-image')
q_dict = pyTEMlib.eds_tools.load_k_factors()
tags = pyTEMlib.eds_tools.quantify_eds(spectrum, q_dict, mask = [])
print(spectrum.metadata['EDS']['GUI'])
print('\n single_atom')
# peaks2, pp2 = pyTEMlib.eds_tools.fit_model(spectrum_atom, use_detector_efficiency=True)
# model2 = pyTEMlib.eds_tools.get_model(spectrum_atom)
# tags2 = pyTEMlib.eds_tools.quantify_eds(spectrum_atom, q_dict, mask = [])
# print(spectrum.metadata['EDS']['GUI'])whole spectrum-image
using L k-factor for Ce
using k-factors for quantification
Al: 55.57 at% 34.34 wt%
Ce: 6.56 at% 21.03 wt%
O : 3.68 at% 1.35 wt%
Cu: 17.75 at% 25.84 wt%
Ni: 4.41 at% 5.93 wt%
Mn: 2.51 at% 3.16 wt%
Zn: 3.90 at% 5.83 wt%
F : 4.93 at% 2.15 wt%
Mg: 0.69 at% 0.38 wt%
excluded from quantification []
{'Al': {'symmetry': 'K-family', 'atom%': np.float64(55.57101162209383), 'weight%': np.float64(34.335477854684974), 'excluded': False, 'k_factor': 0.330728, 'intensity': np.float64(41048.82103182718)}, 'Ce': {'symmetry': 'L-family', 'atom%': np.float64(6.555195911537163), 'weight%': np.float64(21.03364343841367), 'excluded': False, 'k_factor': 0.531928, 'intensity': np.float64(15634.723772788473)}, 'Cu': {'symmetry': 'K-family', 'atom%': np.float64(17.75464745141047), 'weight%': np.float64(25.836220037181008), 'excluded': False, 'k_factor': 0.513247, 'intensity': np.float64(19903.5760208229)}, 'Mn': {'symmetry': 'K-family', 'atom%': np.float64(2.5097844526595905), 'weight%': np.float64(3.157460838620707), 'excluded': False, 'k_factor': 0.424546, 'intensity': np.float64(2940.6394300918255)}, 'Zn': {'symmetry': 'K-family', 'atom%': np.float64(3.8960021929657502), 'weight%': np.float64(5.8330112227936315), 'excluded': False, 'k_factor': 0.542706, 'intensity': np.float64(4249.6850573020665)}, 'Ni': {'symmetry': 'K-family', 'atom%': np.float64(4.408873137760757), 'weight%': np.float64(5.925437410744182), 'excluded': False, 'k_factor': 0.465132, 'intensity': np.float64(5037.009303040206)}, 'Mg': {'symmetry': 'K-family', 'atom%': np.float64(0.6892312380534368), 'weight%': np.float64(0.38360943592475716), 'excluded': False, 'k_factor': 0.327781, 'intensity': np.float64(462.73682487077144)}, 'O': {'symmetry': 'K-family', 'atom%': np.float64(3.683717416374447), 'weight%': np.float64(1.3496423793466454), 'excluded': False, 'k_factor': 0.371908, 'intensity': np.float64(1434.8673162457708)}, 'F': {'symmetry': 'K-family', 'atom%': np.float64(4.93153657714457), 'weight%': np.float64(2.145497382290426), 'excluded': False, 'k_factor': 0.385149, 'intensity': np.float64(2202.5601185034734)}}
single_atom
q_dict = pyTEMlib.eds_tools.load_k_factors()
tags = pyTEMlib.eds_tools.quantify_eds(atom_sum, q_dict, mask = [])
using L k-factor for Ce
using k-factors for quantification
Ni: 3.56 at% 4.54 wt%
Cu: 30.97 at% 42.74 wt%
Ce: 4.52 at% 13.74 wt%
Al: 55.99 at% 32.81 wt%
Zn: 2.96 at% 4.21 wt%
Mg: 0.63 at% 0.33 wt%
Mn: 1.37 at% 1.63 wt%
excluded from quantification []
4.52/1.373.2992700729927003Absorption Correction¶
Lower energy lines will be more affected than higher x-ray lines.
At thin sample location (<50nm) absorption is not significant.
v = spectrum_atom.plot()# ------ Input ----------
thickness_in_nm = 150
# -----------------------
pyTEMlib.eds_tools.apply_absorption_correction(spectrum, thickness_in_nm)
for key, value in spectrum.metadata['EDS']['GUI'].items():
if 'corrected-atom%' in value:
print(f"Element: {key}, Corrected Atom%: {value['corrected-atom%']:.2f}, Corrected Weight%: {value['corrected-weight%']:.2f}")Element: Al, Corrected Atom%: 56.40, Corrected Weight%: 35.34
Element: Ce, Corrected Atom%: 6.34, Corrected Weight%: 20.65
Element: Cu, Corrected Atom%: 17.16, Corrected Weight%: 25.32
Element: Mn, Corrected Atom%: 2.43, Corrected Weight%: 3.10
Element: Zn, Corrected Atom%: 3.77, Corrected Weight%: 5.72
Element: Ni, Corrected Atom%: 4.26, Corrected Weight%: 5.81
Element: Mg, Corrected Atom%: 0.72, Corrected Weight%: 0.40
Element: O, Corrected Atom%: 3.87, Corrected Weight%: 1.44
Element: F, Corrected Atom%: 5.06, Corrected Weight%: 2.23
Find Atoms¶
v = haadf_sum.plot()import skimage
# ------- Input ------
threshold = 1.5 #usally between 0.01 and 0.9 the smaller the more atoms
atom_size = .05 #in nm
# ----------------------
image = haadf_sum/haadf_sum.max()*100
image -= image.min()
scale_x = image.get_image_dims(return_axis=True)[0].slope
#image = image_choice.dataset
# scale_x = pyTEMlib.file_tools.get_slope(image.dim_1)
blobs = None
blobs = skimage.feature.blob_log(image, max_sigma=atom_size/scale_x, threshold=threshold)
fig1, ax = plt.subplots(1, 1,figsize=(8,7), sharex=True, sharey=True)
plt.title("blob detection ")
plt.imshow(image.T, interpolation='nearest',cmap='gray', vmax=np.median(np.array(image))+3*np.std(np.array(image)))
plt.scatter(blobs[:, 0], blobs[:, 1], c='r', s=20, alpha = .5);from tqdm.notebook import trange
neighbor_tree = scipy.spatial.KDTree(blobs[:, :2])
data_array = np.array(dataset)
atom_spectra = np.zeros([len(blobs), dataset.shape[2]])
for x in trange(image.shape[1]):
for y in range(image.shape[0]):
index = neighbor_tree.query([x,y])[1]
atom_spectra[int(index)] += data_array[x,y]
plt.figure()
plt.plot(spectrum.energy_scale, atom_spectra[18], label='atom spectrum1')
plt.plot(spectrum.energy_scale, atom_spectra[57], label='atom spectrum2')
plt.plot(spectrum.energy_scale, atom_spectra[38], label='atom spectrum3')
plt.legend()voronoi =scipy.spatial.Voronoi(blobs[:, :2])
plt.figure()
plt.imshow(image.T)
scipy.spatial.voronoi_plot_2d(voronoi, plt.gca(), show_vertices=False, line_colors='orange', line_width=1, line_alpha=0.6, point_size=2);Summary¶
The spectrum is modeled completely with background and characteristic peak-families.
Either
k-factors in a file (here from Spectra300) or
Bothe-Salvat cross-sections
are used for quantification.
Appendix¶
Background¶
The determined background used for the model-based quantification is based on the detector effciency.
Note:
The detector efficiency is also used for the quantification model.